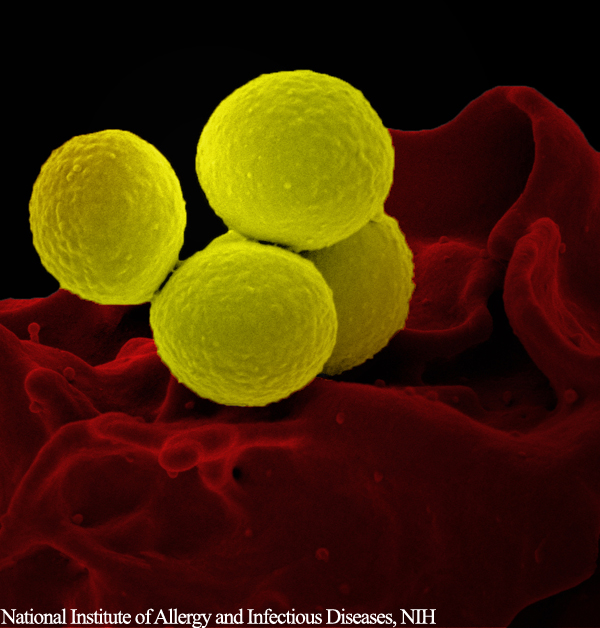
Researchers from Arizona State University have developed a technique to identify antibiotic resistant bacteria, and differentiate them from their antibiotic sensitive counterparts. Using microfluidic technology, the scientists were able to sort the bacteria present in a sample in just a few minutes.
Antibiotic resistance is an ever-expanding issue in healthcare, which results in fewer treatment options for patients, prolonged illnesses, and increased mortality rates. Currently, tests used to identify whether or not a person’s infection will be resistant to treatment with conventional antibiotics takes multiple days. In the case of a potentially life-threatening infection, time is a key factor.
Professor Mark Hayes and his lab in the Department of Chemistry and Biochemistry at Arizona State University, developed the microfluidic technology that utilizes a microscopic electric field gradient, and very small sample volumes, to identify the resistant and susceptible strains of Staphylococcus epidermidis.
Development of this new technique is evidence of a shift in the way bacterial infections are handled. Following decades of an all-out war on bacteria – from antibacterial soaps, detergents and hand sanitizers, to the overuse of prescription antibiotics – researchers are approaching the issue in a subtler way. Scientists are now focusing on understanding what role bacteria play in our bodies and our environment, and are intent on identifying the pathogenic strains from the beneficial.
Hayes and his team fabricated a glass and silicone polymer chip, containing microscopic channels through which bacterial cells can pass. These microchannels feature sawtooth-shaped grooves, which enable the researchers to sort bacterial species using electrophoresis and the bacteria’s unique electrochemical signature.
The electrochemical characteristics of a strain can vary based on whether the bacteria is resistant or susceptible to antibiotic treatment. Paul V. Jones and Shannon Hilton, graduate students in the Hayes lab, used this technique to separate Gentamycin-resistant and susceptible bacteria of the same species.
“The important thing for the patient and the doctor is getting the right answer right away. By advancing a fundamental area of science, we are able to create an entirely new way to differentiate these microorganisms,” said Hayes. “And it turns out that we have a core mechanism that could be integrated with smartphones and be widely, and cheaply, distributed. Frankly, we only hoped that this strategy could work this well, even though our theoretical calculations suggest that it could.”
The researchers used unique fluorescent dyes to visually tell the two types of Staphylococcus epidermidis apart. After injecting the bacteria into the microchannels and applying the voltage, the dielectrophoretic force allowed some bacteria to migrate further, based on their phenotype.
This simple, rapid method for detecting difficult-to-treat bacterial strains, has strong therapeutic potential. These results will be influential in both early detection of antibiotic infection, as well as in the development of novel antibiotic targets.
Sources:
- ASU team develops quick way to determine bacteria’s antibiotic resistance – https://asunews.asu.edu/20150901-bacteria-resistance-research
- Jones, P., Huey, S., Davis, P., McLemore, R., McLaren, A., and Hayes, M. (2015). Biophysical separation of Staphylococcus epidermidis strains based on antibiotic resistance. Analyst. 140, 5152-5161.










Join or login to leave a comment
JOIN LOGIN