A Comparison to TNM Staging in Bladder Cancer
Machine and deep learning, key components of image analysis, enable the analysis of complex visual patterns. In this free webinar, the featured speakers will tackle the challenges of quantitative pathology and more specifically of immunofluorescence (IF) imaging, such as the variability of observed biological patterns, the inter-slide and intra-slide variability of staining and the presence of non-specific staining. The automated detection of nuclei and epitheliumregions allows users to compute a set of features related to the immune contexture (e.g. the ratio of CD8 positive cells in the non-epithelium regions) and to the tumor morphology (e.g. the number of tumor buds in tumour core). Based on these features, the time‐to‐event data of bladder cancer patients were modelled using survival decision trees, and immune-related and tumor bud-related models were derived, both of which may provide prognostic value for survival and better risk stratification of bladder cancer patients than TNM staging.
Motivation – Muscle-invasive bladder cancer (MIBC) is a highly aggressive disease whose clinical reporting is based on TNM staging. Despite recent research into novel treatment and surgical strategies the mortality rates andprognoses of MIBC patients have remained immutable over the past 30 years. In addition to inter- and intra-reporter variability, TNM staging may not adequately encompass the complex and dynamic behavior of the disease. There is, therefore, a need to substantially improve patient stratification and earlier definitive treatment for high-risk MIBC patients. An analysis of the tumor morphology and of the immune contexture may hold important pathological information, which could allow for better risk stratification of patients with MIBC than current clinical guidelines based on TNM staging.
Quantitative pathology – An image analysis solution based on machine learning and deep learning was developed to automatically quantify cell populations across IF labelled whole slide images from MIBC patients with known survival data. Cell centers and epithelium regions are automatically detected using novel algorithms based on visual context random forest and convolutional neural networks respectively. By enabling the automatic definition of high-dimensional decisions based on color and hierarchical texture information, the employed algorithms can tackle inherent complexity of the scene (e.g. large inter- and intra-sample heterogeneity, presence of tissue artefacts and necrotic debris).
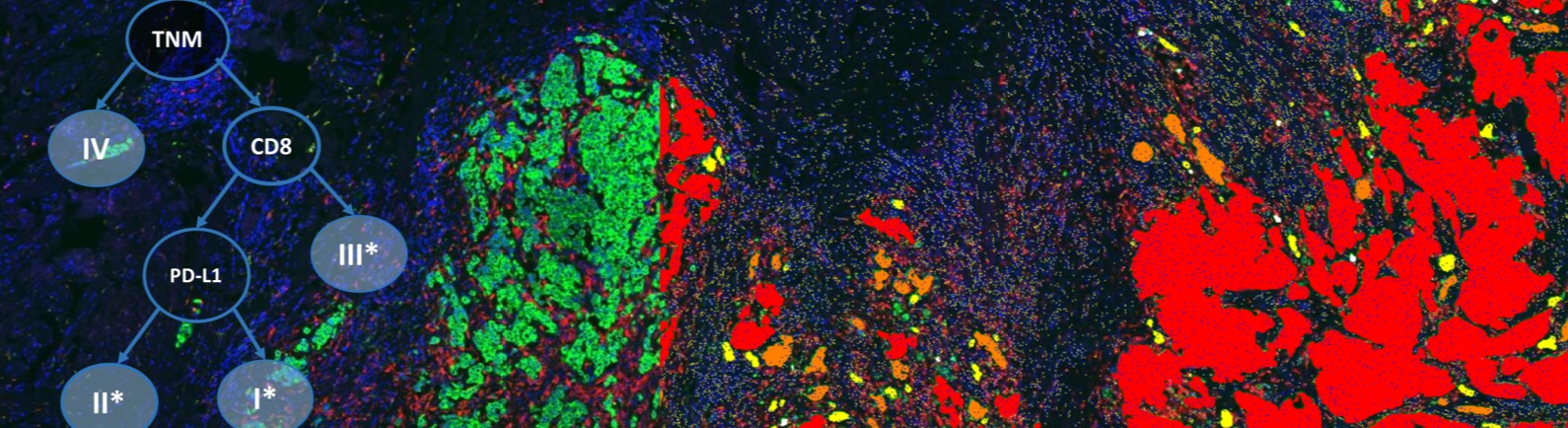
Immune contexture – Following the classification of the detected cells into three subtypes based on the CD3, CD8 and PD-L1 channels, we calculate for each patient the proportions of the different detected cell subtypes within the detected epithelium and non-epithelium regions. Using these proportion features, patients were then automatically split into different survival groups using a cross-validated survival decision tree. The resulting decision tree stratifies MIBC patients into three immune-related groups (I*, II*, III*) utilizing the proportion of CD8(+) cells in the non-epithelium regions as a first decision and the proportion of PD-L1(+) cells across the whole tissue as a second decision within the sub-group of patients with high proportion of CD8(+) cells. The final grouping is obtained by substituting the TNM stages I, II and III by the three identified immune groups I*, II*, and III* on non-metastatic patients. Testing for survival curve differences suggests that the proposed grouping based on immune profiling provides prognostic value for survival and better risk stratification of patients than TNM stage.
Tumor budding – In addition to automatically quantifying the immune contexture, the proposed image analysis solution enables the detection and quantification of small cancer cell clusters — the tumor buds — which have been suggested to reflect an early step in progression towards metastasis. Splitting non-metastatic MIBC patients of stage II and III into low and high tumor budding groups seems to confer prognostic significance in risk stratification of MIBC patients, based on tumor budding quantification. his is the first evidence of tumor budding holding prognostic significance in MIBC.
Conclusion – While the joint impact of tumor buds and immune context for survival is still under investigation, this study shows the value of quantifying immune context and tumor morphology using quantitative pathology on immunofluorescence images. More generally, and upon confirmation of the findings on multi-site independent validation cohorts, current clinical guidelines based on TNM could be refined to encompass the proposed immune and morphological quantification of the disease.
Speakers

Dr. Nicolas Brieu, Principal Research Scientist, Definiens
Nicolas Brieu received his PhD from the Technical University Munich (TUM) with a special focus on computer vision for medical imaging. As a principal research scientist at Definiens, Nicolas leads the research and development of novel algorithms to automate the analysis of digital pathology images and the identification of immune-related prognostic factors. Nicolas has published over 20 papers in the area of medical image, machine learning, deep learning and data mining.

Dr. Peter D. Caie, Senior Research Fellow, St. Andrew University, UK
Dr. Caie is a Senior Research Fellow at the University of St Andrews and is Principal Investigator at the QUAntitative and Digital (QUAD) pathology group. His work concentrates on applying digital pathology, complex image analysis, integrative big data, multi-omics, and artificial intelligence to answer clinically transferable research questions. He submitted his Ph.D. in Digital and Quantitative Pathology at the University of Edinburgh while building a Scottish wide Quantitative and Systems Pathology collaborative group spanning both clinical and academic settings. Previously, Dr. Caie attained an M.Res in Medical biochemistry and a B.Sc in Molecular and Cellular biology, both from the University of Glasgow. Dr. Caie also worked in Industry with AstraZeneca for nine years where he developed high content biology assays for in vitro drug discovery.
Who Should Attend?
This webinar will be suitable for:
- Translational Scientists
- VPs of Oncology
- Directors of Immuno-Oncology programs
- Directors of Companion Diagnostics
- Directors of Profiling
- Clinical Trial Managers/Directors/VP
What You Will Learn
In this free webinar, participants will learn about:
- How challenges in the analysis of IF images are overcome with machine learning and in particular deep learning, therefore enabling the automated quantification of immune contexture and tumor morphology
- How survival decision trees enable the generation of easy-to-interpret models to stratify patients based on immune-related and morphological features and on censored survival data.
- How these models result in the identification of sub-groups of patients with prognostic value
- Combined use of machine learning (visual context random forest) for cell detection and of deep learning (convolutional neural networks) for epithelium region segmentation
Xtalks Partner
Definiens
We improve patient lives by unlocking the tissue phenome.
In oncology, therapeutic strategies have shifted from a direct assault on cancer cells to recruiting the immune system for that purpose. Our mission is to accelerate breakthroughs for this approach by helping scientists leverage Tissue Phenomics to deepen understanding of disease biology and immune system mechanisms, to bring multi-omics data into a cancer-relevant context, and to facilitate the translation of new insights into novel therapies and treatment strategies. Our vision is to create unique patient profiles for an individualized standard of care, where patients experience fewer side effects and live longer.
Definiens’ Tissue Phenomics approach was awarded the 2013 Frost and Sullivan Company of the Year Award for Global Tissue Diagnostics and Pathology Imaging. For more information, please visit: www.definiens.com.
You Must Login To Register for this Free Webinar
Already have an account? LOGIN HERE. If you don’t have an account you need to create a free account.
Create Account