How Bioinformatics Contributes to In Silico Manufacturability Assessments
Monoclonal antibodies (mAbs) and novel antibodies can display good specificity and affinity for their targets, but their physical and chemical instability can have a detrimental impact on their capacity to be manufactured effectively. Post-translational modifications and charge heterogeneity enhance the possibility of immunogenicity, while aggregation decreases protein function and increases concerns about an immune response.
The ability to identify liabilities of a molecule is extremely important for process, analytical method, and formulation development. At FUJIFILM Biotechnologies, we understand the importance of de-risking candidate selection, accelerating time to clinic and that bioinformatics can play a pivotal role, so we start these conversations with our clients from the very outset.
Our established in silico workflows cater for the diverse range of molecules in our development and manufacturing pipelines, which can span from simple proteins to molecules with complex domain architectures such as fusion proteins and bispecific monoclonal antibodies (Figure 1A). Using available or derived molecular descriptors, we can screen and rank multiple candidates to provide our partners with data-driven lead candidate selection options (Figure 1B).
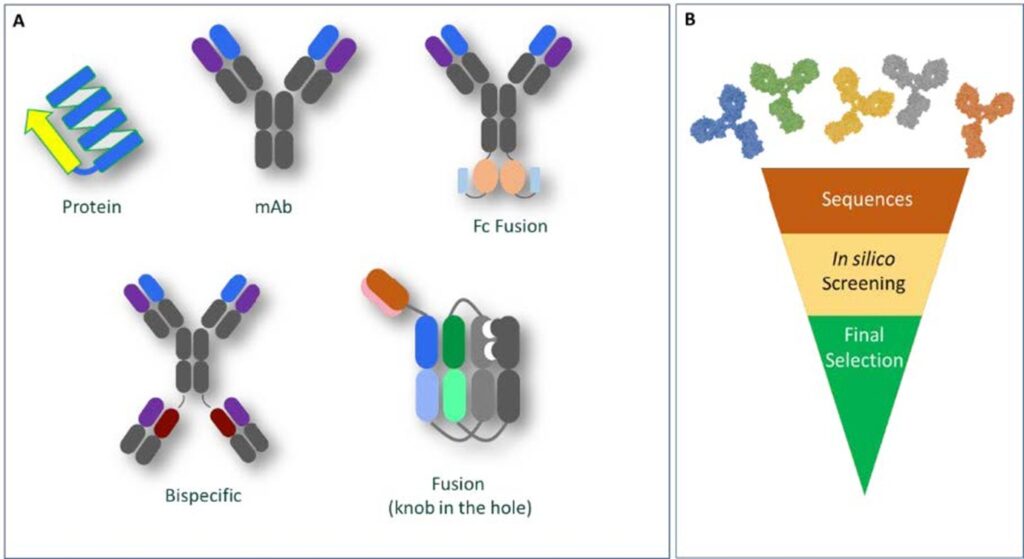
Figure 1A: Diversity of molecules assessed using bioinformatics; Figure 1B: Representation of lead candidate selection ‘funnel.’
Our Approach to Bioinformatics
Our global approach is supported by four pillars, as shown below in Figure 2. These four pillars include: derivation of sequence-based features, building structural 3D models, exploring molecular flexibility, and analysis of the complex multi-dimensional data for a better understanding of the molecule and assessing its manufacturability risks.
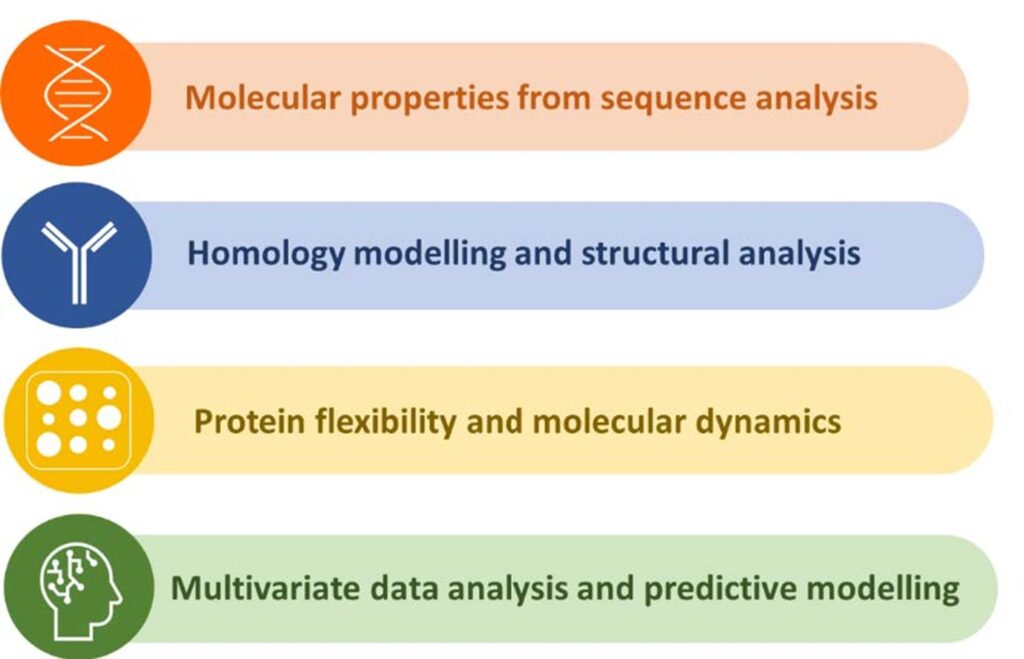
Figure 2: Core pillars of bioinformatics at FUJIFILM Biotechnologies.
Optimized Resource for Biological Information and Toolkits (ORBIT) is an in-house-developed, self-serviced bioinformatics software database and application platform. ORBIT serves as a centralized database for biologics information while also hosting calculation and prediction algorithms. ORBIT calculates physicochemical properties and uses integrated machine learning and advanced multivariate data analysis to predict the quality attributes and assess manufacturability risk for biological molecules. Built-in interactive tools and visualizations assist our scientists in understanding discrete liabilities prior to the molecule’s entry into Process Development workflows.
Molecular structure and spatial orientation determine the characteristics of the molecule and its efficacy. At FUJIFILM Biotechnologies, a sophisticated modelling software, Molecular Operating Environment (MOE), is used to build a 3D homology model of the molecule to derive ensemble-based structural properties for risk assessment. Use of MOE contributes to the manufacturability assessment of a molecule by calculating biophysical properties, visual analysis, atomistic detail description, and surface patch analysis. Chemical liabilities such as Asn deamidation, Asp isomerization, Lys glycation and Met oxidation are analyzed to identify the potential risk.
In nature, all molecules are dynamic, and protein dynamics plays a crucial role in the structure-function of the molecule. At FUJIFILM Biotechnologies, we explore protein dynamics using GROMACS (GROningen MAchine for Chemical Simulations), an open-source software for molecular dynamics simulations. This state-of-the-art tool is used to determine the conformational phase space of the biologic to derive time-dependent evolution of protein structures. Exploring the free energy landscape and calculation of thermodynamic properties of these molecules helps to provide a deeper understanding of dynamic behavior, which is useful during process development stages.
Overall, bioinformatics generates a vast amount of data. Starting from exploratory data analysis, leading to multivariate data analysis and advanced machine learning methods, we can develop novel algorithms and predict critical properties for our clients’ molecules to identify manufacturability risks. Of equal importance to the data itself is the way that it is interpreted. We use commercial software (such as JMP) and open-source software (such as R) to analyze data, extract insights and derive meaningful conclusions to help our clients make faster and better decisions.
Methods and Results
To illustrate FUJIFILM Biotechnologies’ approach to lead candidate screening, in silico manufacturability assessment factors outlined in Table 1 were applied to four candidate mAbs (A-D).
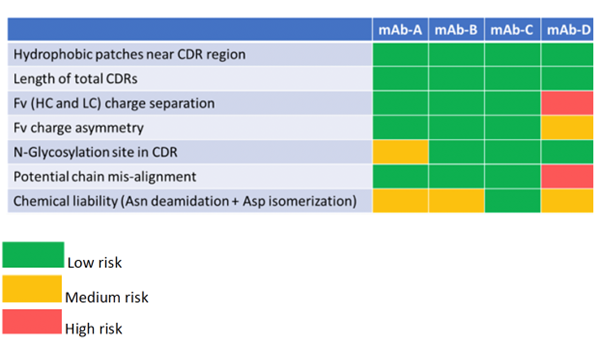
Table 1: In silico risk assessment of mAb candidates A-D.
The first 3 factors (hydrophobic patches near CDR region, length of total CDRs and Fv [HC and LC] charge separation) are considered as critical early-assessment parameters1 compared to lower risks, such as additional glycosylation sites, chemical liabilities, Fv charge asymmetry, and potential chain misalignment. At FUJIFILM Biotechnologies, all factors were evaluated using in silico methods developed in-house.
mAb-D was identified as having two high and two medium risks. mAb-A, on the other hand, has only 2 medium risks: an additional glycosylation site in the CDR and two chemical liabilities in the Fv, which might introduce charge heterogeneity in the molecule. One medium risk parameter (chemical liability) was identified in mAb-B. Overall, mAb-C was the lowest risk (and was also observed as having the lowest number of hydrophobic patches compared to the others).
Based on the light chain subclass, mAb expressibility can also be classified. It was observed that the kappa-III chain is more expressive than other kappa chains, followed by the lambda chains. Since mAb-B contained a kappa-III light chain, while mAb-C contained a kappa-I light chain, mAb-B may be a better choice when considering expressibility.
Conclusion
As expressibility needs to be balanced with other risk factors, we proposed both mAb-B and mAb-C as potential lead candidates. Out of two leads, mAb-C was taken forward for cell line development and the clonal cell line produced 3.6 g/L of product concentration in an AMBR15® screen.
Clarified harvest supernatant samples were partially purified by protein A chromatography, analysis by size exclusion UPLC revealed <3% of high molecular weight species and >93% of monomeric species, and >94% total IgG by CE-SDS under reducing conditions. In our experience, these data from a partially purified sample provide sufficient confidence that subsequent polishing chromatography steps will be capable of generating DS of higher quality, typically >98% monomeric full-length molecule by size exclusion UPLC.
To assimilate and interpret data, experience matters. Our clients have credited our staff and bioinformatics package as being one of the most comprehensive and informative in the industry.
References
1) MAbs,2021 Jan-Dec;13(1):1981805. doi: 10.1080/19420862.2021.1981805.
About the Author
 Arghya Barman joined FUJIFILM Biotechnologies in 2018 to lead our efforts and develop in-house computational applications in Bioinformatics. He brought with him a wealth of experience from post-doctoral studies in computational structural biology and as a Bioinformatician at INVISTA textiles, where he designed industrial biocatalysts using state-of-the art in silico methods. His role in Data Science also includes providing data-driven insights for challenging scientific problems using advanced data analysis and machine learning algorithms. He received his PhD in Computational Chemistry from the University of Miami in the United States.
Arghya Barman joined FUJIFILM Biotechnologies in 2018 to lead our efforts and develop in-house computational applications in Bioinformatics. He brought with him a wealth of experience from post-doctoral studies in computational structural biology and as a Bioinformatician at INVISTA textiles, where he designed industrial biocatalysts using state-of-the art in silico methods. His role in Data Science also includes providing data-driven insights for challenging scientific problems using advanced data analysis and machine learning algorithms. He received his PhD in Computational Chemistry from the University of Miami in the United States.
About FUJIFILM Biotechnologies
FUJIFILM Biotechnologies, a subsidiary of FUJIFILM Corporation, is a world-leading contract development and manufacturing organization (CDMO) for biologics, vaccines and advanced therapies. With over 30 years of experience, the Company specializes in developing and manufacturing biopharmaceuticals using microbial, mammalian, and host/virus systems. With over 4,800 employees, FUJIFILM Biotechnologies operates a fully integrated, kojoX™ global network with major facilities in the United States, the United Kingdom, and Denmark, with a new site in Holly Springs, North Carolina, USA. The Company’s kojoX™ manufacturing network ensures supply chain agility for its customers through modular facilities and standardized processes for seamless scaling and technology transfers. FUJIFILM Biotechnologies offers comprehensive services, ranging from proprietary cell line development, to process and analytical development, and through to clinical and commercial manufacturing. For more information, go to: fujifilmbiotechnologies.fujifilm.com or contact the FUJIFILM Biotechnologies team.



Join or login to leave a comment
JOIN LOGIN